There are relatively few options in R for comparing matched pairs in two groups with left-censored data. And while NADA2::cen_signedranktest() is an excellent tool, I wrote the following function as another. It implements the Paired Prentice-Wilcoxon test, as described in Helsel (2012). I should also acknowledge the USGS’ orphaned package smwrQW (Lorenz 2017) for its version of the same test.
library("tidyverse")
library("survival")
ppw_test <- function(
x, # numeric
y, # numeric
x_cen, # logical
y_cen, # logical
alternative = "two.sided", # either "two.sided", "greater" (x > y), or "less" (x < y)
flip = TRUE
) {
if(length(x) != length(y))
stop("Lengths of x and y must be the same for paired data.")
if(any(c(x, y) < 0))
stop("Negative values in x or y.")
valid_alternative <- pmatch(alternative, c("two.sided", "greater", "less"))
if(is.na(valid_alternative))
stop('Invalid choice for alternative, must match "two.sided", "greater", or "less"')
test_input <- tibble(x_val = x, y_val = y, x_cen, y_cen) %>%
na.omit() %>%
rowid_to_column() %>%
pivot_longer(
cols = -rowid,
names_to = c("group", ".value"),
names_pattern = "(.+)_(.+)"
) %>%
mutate(
cen = as.numeric(!cen), # 0 is censored, 1 is observed
# flip data so that smallest observation becomes longest "survival time":
# N.B., this rounds the flipped data to 6 decimal places
flipped = if(flip) {max(val) + 1 - val} else val,
flipped = round(flipped, 6)
) %>%
left_join(
# estimate survival function:
survival::survfit(survival::Surv(flipped, cen) ~ 1, data = .) %>%
broom::tidy() %>%
mutate(time = round(time, 6)),
by = c("flipped" = "time")
) %>%
mutate(score = if_else(cen == 1, 1 - 2 * estimate, 1 - estimate)) %>%
pivot_wider(id_cols = rowid, names_from = group, values_from = score) %>%
mutate(d = x - y) %>%
summarize(
z_ppw = sum(d) / sqrt(sum(d ^ 2)),
p_val = if(alternative == "two.sided") {2 * pnorm(abs(z_ppw), lower.tail = FALSE)} else
if(alternative == "greater") {pnorm(-z_ppw, lower.tail = FALSE)} else
if(alternative == "less") {pnorm(z_ppw, lower.tail = FALSE)} else
"alternative hypothesis is invalid"
)
list("statistic" = test_input$z_ppw, "p.value" = test_input$p_val)
}
The ppw_test() function works like this:
withr::with_seed(450, { # generate two random normal variables, with left-censoring:
tibble(
x = rnorm(10, 3, 1),
y = rnorm(10, 3, 1),
x_cen = x < 2,
y_cen = y < 2
)
}) %>%
with(ppw_test(x, y, x_cen, y_cen))
$statistic
[1] -0.5628925
$p.value
[1] 0.5735081It also does one-sided tests:
withr::with_seed(23, {
tibble(
x = rnorm(10, 10, 1),
y = rnorm(10, 5, 1),
x_cen = x < 10,
y_cen = y < 5
)
}) %>%
with(ppw_test(x, y, x_cen, y_cen, alternative = "greater"))
$statistic
[1] -2.918231
$p.value
[1] 0.001760118The following code tests that ppw_test() gives the expected result when applied to the data in Table 9.7 of Helsel (2012). First, here are the data:
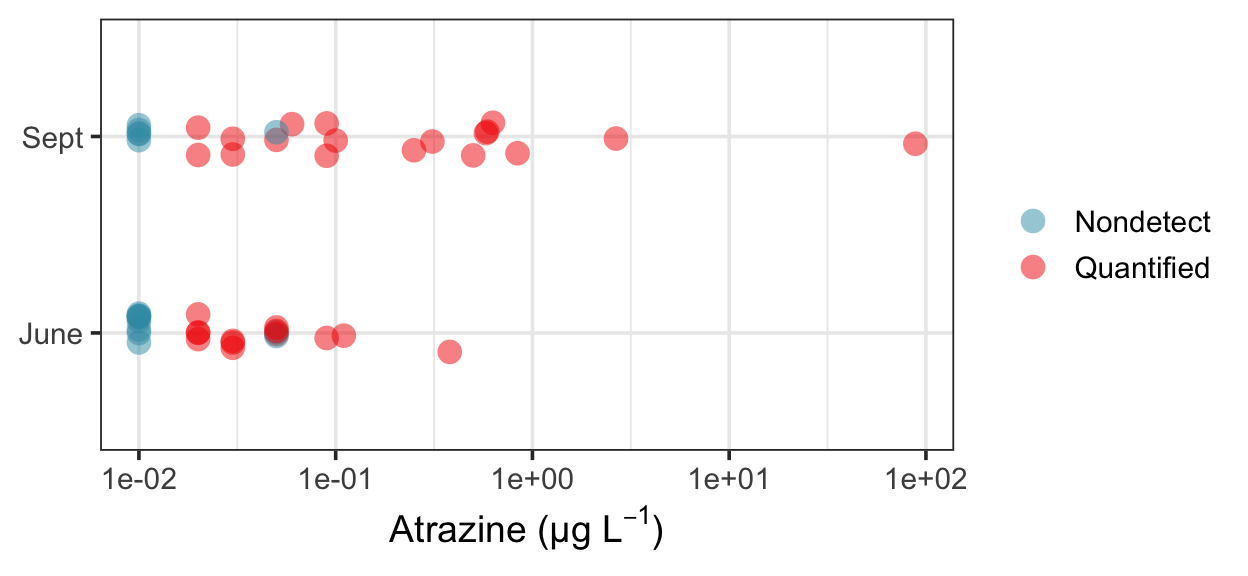
And here are the test results. These are close to the values in Helsel (2012), but not exactly the same. I suspect there are a few typos in the table, which may have something to do with it. For example, while most of the scores calculated by ppw_test() are consistent with those reported in the table, line 11, column 3 list a score of 0.55 for the second-highest value in column 1, while ppw_test() calculates a score of -0.54, much closer to the value of -0.67 corresponding to the largest value in column 1. There is a similar problem on line 12.
helsel %>%
mutate(
june_cen = str_detect(june, "<"),
sept_cen = str_detect(september, "<"),
) %>%
mutate_if(is.character, ~ as.numeric(str_remove(.x, "<"))) %>%
with(ppw_test(june, september, june_cen, sept_cen, "less"))
$statistic
[1] 3.012394
$p.value
[1] 0.001295979Helsel, Dennis R. 2012. Statistics for Censored Environmental Data Using Minitab and R. 2nd ed. Wiley Series in Statistics in Practice. Hoboken, N.J: Wiley.
Lorenz, Dave. 2017. “SmwrQW–an R Package for Managing and Analyzing Water-Quality Data, Version 0.7.9.” Open File Report. U.S. Geological Survey.